Intelligence
links: reference: https://www.sciencedirect.com/journal/intelligence/ Don’t sleep on this http://www.iqscorner.com/ https://en.wikipedia.org/wiki/Outline_of_human_intelligence https://unexaminedglitch.com/f/why-the-mouse-runs-the-lab-and-the-psychologist-is-in-the-maze https://socialsci.libretexts.org/Bookshelves/Psychology/Biological_Psychology/Biopsychology_(OERI)_-_DRAFT_for_Review/14%3A_Intelligence_and_Cognition this seems to be a pretty high-brow book despite having ‘psychology’ in the name. Some good chapters.
- https://www.lesswrong.com/posts/cLr6TJj2qRrBa3Wmu/intelligence-enhancement-monthly-thread-13-oct-2023
- The Biointelligence Explosion (David Pearce 2012)
- The Computational Boundary of a “Self”: Developmental Bioelectricity Drives Multicellularity and Scale-Free Cognition (Levin 2019)
- https://www.reddit.com/r/cognitiveTesting/comments/146fmpr/comprehensive_online_resources_list/ 10-25-2021
Intelligence #
-
Savantism often has damage to the left anterior Temporal Lobe; it can be artifically replicated by disabling via TMS
-
Apparently patients who have undergone hemispherectomies experience little differences, including IQ. Not to be confused with split-brain, in which the corpus callosum is cut and the hemispheres are divorced. R
-
Macronephaly: greater brain size; associated with autism; apparently does not increase intelligence
-
Brain volume: r=~.4
-
External head size: r=~.2
-
Frontal, temporal, parietal lobe and hippocampus volume: r=~.25
-
Non-verbal g and corpus callosum size: r=~.47
-
Overall grey and white matter: r=~.3
-
Absolute regional glucose metabolism: r=~-0.48 - -0.84
-
Neuron action potential speed: r=.37 Intelligence: Knowns and Unknowns
-
Cortical gyrification/brain surface area? Up to 85% BSA variance can be explained by genes.
-
Possibly more upper-range Alpha waves, and a smaller extent of cortical activation. R
-
Positive correlation with mid-brain SERT availability and IQ. R
-
Fluid intelligence and the locus coeruleus–norepinephrine system
-
Intelligence IS Cognitive Flexibility: Why Multilevel Models of Within-Individual Processes Are Needed to Realise This #
-
- Correlated with fewer connections??
-
Graph lesion-deficit mapping of fluid intelligence #
-
https://pumpkinperson.com/2020/03/31/the-true-distribution-of-intelligence/ * So even though IQ 119 is about twice as high as IQ 58, the difference in actual problem solving speed is about an order of magnitude*
-
Testing: Darius: Problems in Gentle Slopes 2, Alchemist Test, and also, there two lesser known tests which are very good isolaters of strictly pattern rec one of them is called SEQSI and the other is NSL36. https://iq-tests-for-the-high-range.com/tests.html
Genetic Associations #
- https://research.vu.nl/en/persons/natalia-goriounova/publications/ THIS is a researcher worth paying attention to.
- Genetic Mechanisms Underlying the Evolution of Connectivity in the Human Cortex
- https://www.ebi.ac.uk/gwas/search?query=Intelligence
-
The impact of rare protein coding genetic variation on adult cognitive function skimmed. Uhh I think I’m missing something but these might be bad genes or something. Confusing shit, you know how it is.
https://twitter.com/AlexTISYoung/status/1692164275689668779
- Commentary: Discovering genes that affect cognitive ability (Aug 2023)
- Evidence that rare and common variants overlap in association signals and contribute additively to cognitive function. Our study introduces the relevance of rare coding variants for cognitive function and unveils high-impact monogenic contributions to how cognitive function is distributed in the normal adult population.
- Associated with adult cognitive function through rare coding variants with large effects: (none of these are in the other GWAS!)
- ADGRB2
- KDM5B
- GIGYF1
- ANKRD12
- SLC8A1
- RC3H2
- CACNA1A
- BCAS3
- We also identified five putative cognitive function genes (faisle discovery rate of Q < 5%): NDUFA6, ARHGEF7, C11orf94, KIF26A and MAP1A
- So, n=485k. They had 3 ‘phenotypes’: educational attainment (years of schooling), reaction time, and verbal-numerical reasoning.
- [GWAS of 19,629 individuals identifies novel genetic variants for regional brain volumes and refines their genetic co-architecture with cognitive and mental health traits (Zhao et al. 2019)]
-
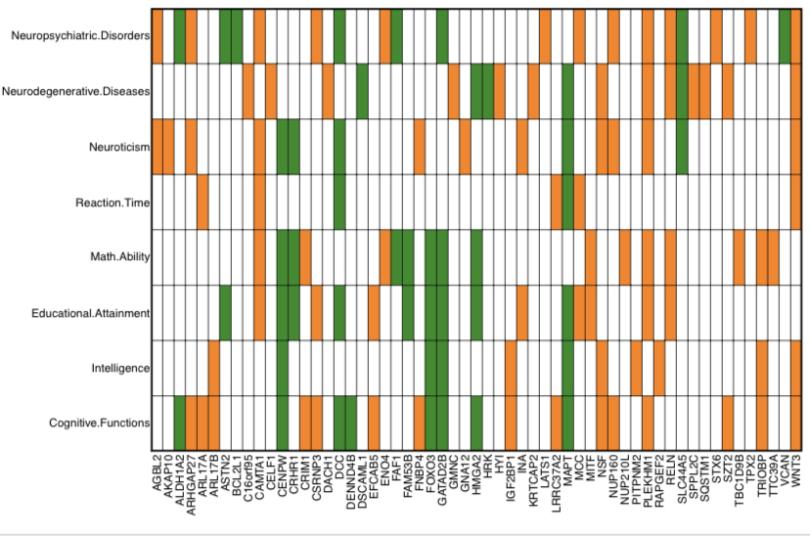 I don’t know any of these besides like FOXO3 and IGF2-BP
I don’t know any of these besides like FOXO3 and IGF2-BP
-
Biological annotation of genetic loci associated with intelligence in a meta-analysis of 87,740 individuals (Coleman et al. 2018/9) #
- Builds upon the two following studies on this page.
- Genes with tissue specificity:
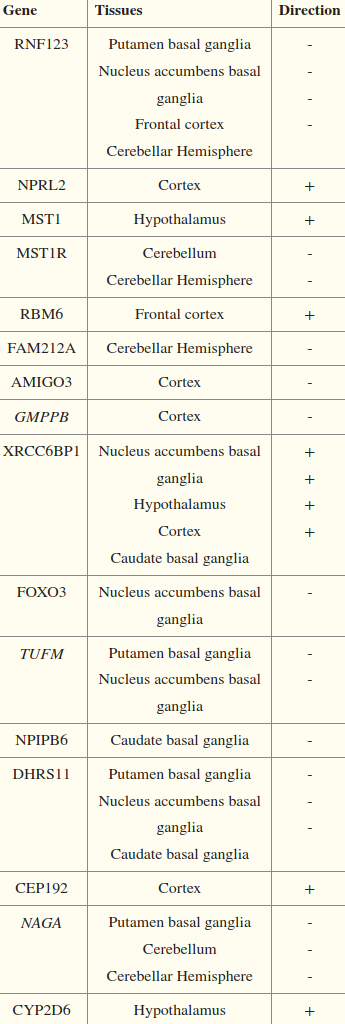
- NPLR2: Nitrogen permease regulator 2-like protein. Tumor suppressor.
- MST1(R): Macrophage-stimulating protein
- RMB6: RNA Binding Motfin/Protein 6
- FAM212A: Family with sequence similarity 212 member A
- AMIGO3: Adhesion molecule with Immunoglobin-like domain 3.
- Predicted to act upstream/within synapse assembly, especially during development.
- GMPPB: Mannose-1-phosphate guanyltransferase beta.
- Converts Mannose-1-phosphate + GTP -> GDP-mannose.
- XRCC6BP1: AKA ATP23. A metallopeptidase.
- FOXO3 negatively correlated in the basal ganglia.
- T (C/C)
- TUFM:
- NPIPB6: Nuclear pore complex interacting protein family member B6
- DHRS11: Dehydrogenase/Reductase 11. Catalyzes conversion of 17-keto steroids into 17β-hydroxyl metabolites. Like it’s 3β-HSD?
- CEP192: Centrosomal protein of 192 kDa. Regulates centrosome maturation/duplication and all kinds of stuff for Microtubule formation/nucleation.
- CYP2D6 positively correlated in the hypothalamus.
-
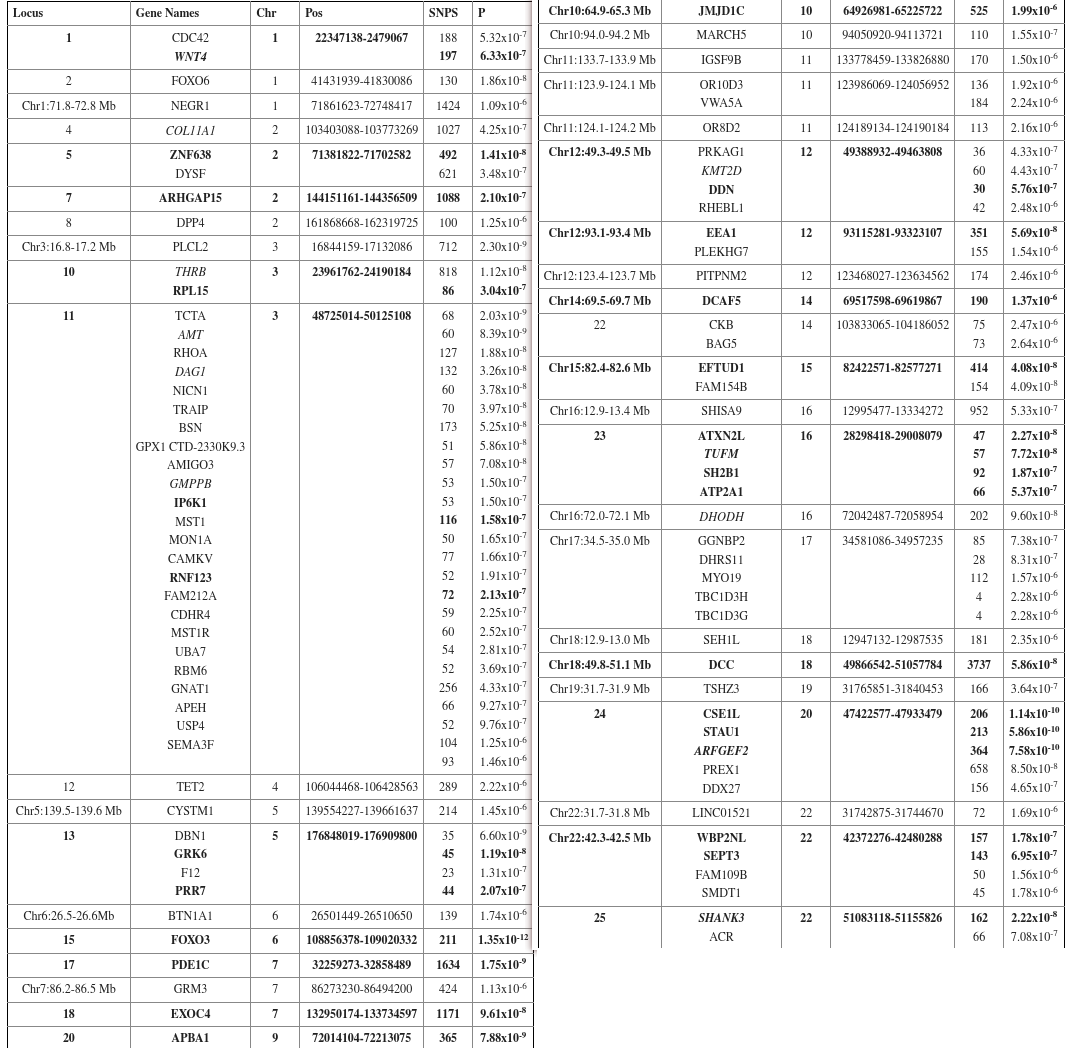
- Bold = seen in the above study. Convenient
- FOXO6
Genome-wide association meta-analysis of 78,308 individuals identifies new loci and genes influencing human intelligence (Sniekers et al. 2017) #
- I got 17th percentile on this according to nebula.
- 13 different cohorts. 18 significant loci, and 336 SNPs (I don’t know how to get to the Supplementary Table).
-
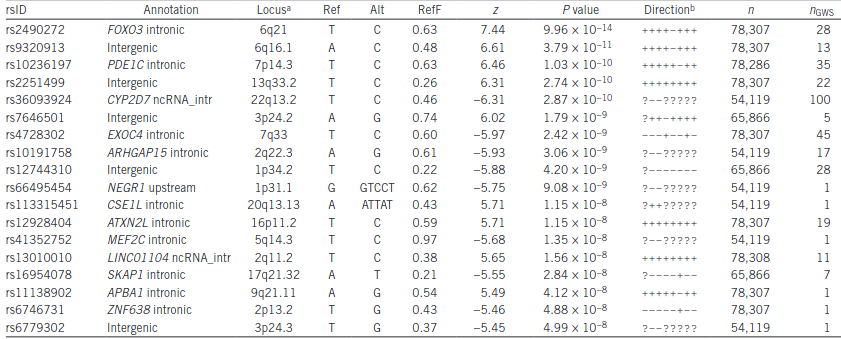
- The direction goes off absolute value. As shown below, it depends on the gene whether activity is increased or decreased on the SNP. ZNF638 seems to have a net positive correlation, thus the SNP must be a loss-of-function.
-
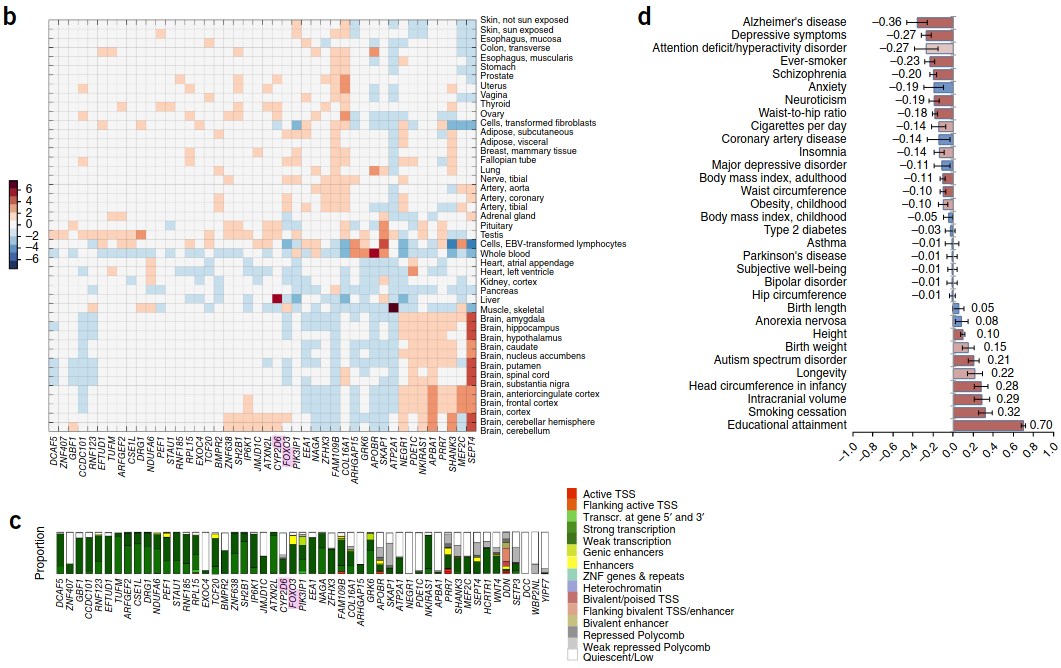 moar genes!
moar genes!
- CSE1L
- EXOC4
- CYP2D6 is located near two P450 ‘psuedogenes’: CYP2d7P/CYP2D8P
- WBP2NL: Part of oocyte fertilization
- APBA1: Member of X11. Stabilizes APP and inhibits production of proteolytic fragments. Putative vesicular trafficking protein.
- STP3: Member of Septin GTPase family. Unkown function but upregulated by retinoic acid in a human teratocarcinoma cell line
- NAGA: N-acetylgalactosinamidase (cleaves NAG moieties from glycoconjugates). Mutations are the cause of Schindler diseases type I/II (aka Kanzaki disease)
- STAU1: Staufen. dsRNA-binding protein. Contains a microtubule-binding domain similar to MAP1B and binds to Tubulin. Associates with the RER, implicating this protein in mRNA transport down microtubules to the RER.
- NDUFA6: Related to ATP, UCP, metabolism, and NADH dehydrogenase activity.
- DCAF5: Associated with leiomyoma
- EFTUD1: Related to Ribosome biogenesis
- DDN (Dendrin): Related to DNA polymerase II core promoter proximal region sequence-specific DNA binding (yeah I don’t know if there’s an acronym for that)
- ZNF407: Unknown Zinc finger protein. ZNF638: Nucleoplasmic protein.
- PDE1C
- T (T/C)
- RPL15/RPL10: Ribosomal protein.
- ATXN2L: Ataxin type 2-related protein.
- SH2B1
- NKIRAS1: NF-κB-inhibitor interacting ras like 1. GTPase.
- TUFM (Tu translation elongation factor): Participates in mitochondrial protein translation. Mutations associated with decline in oxidative phosphorylation and resulting lactic acidosis.
- BMPR2 (Bone morphogenic protein receptor).
- ATP2A1: A SERCA Calcium ATPase
- JMJD1C: Jumonji domain. Interacts with thyroid hormone receptors.
- SHANK3
- ARFGEF2: ADP-ribosylation factor (ARF)
- GRK6
- RNF123: Ring Finger/E3 ubiquitin-protein ligase
- YIPF7 (Yip1 domain member 7)
- GBF1: Guanine nucleotide exchange factor. Localized to the golgi. Activates ADP ribosylation factor 1.
- PEF1: Calcium-binding protein belonging to the penta-EF-hand protein family. Forms a heterodimer with the programmed cell death 6 gene product.
- COL16A1: Type XVI collagen alpha chain.
- DCC: Netrin 1 receptor. Interacts with Src and FAK to mediate axon attraction. Partially locazlies to Lipid Rafts and induces apoptosis in the absence of a ligand. Tumor suppressor.
- PRR7 (Proline rich 7 (synaptic))
- CCDC101: Subunit of ADA2A and SPT3, histone acetyltransferase complexes.
- ARHGAP15: A Rho-regulated GAP.
- SEPT4: Member of septin family of nucleotide binding proteins.
- ZFHX3: Regulates myogenic and nueronal differentiation. Negatively regulates c-Myb and transactivates p21CIP1/p21-1A.
- EEA1 (early endosome antigen 1)
- WNT4
- DRG1 (Developmentally-regulated GTP binding protein 1)
- IP6K1: Inositol phosphokinase 1.
- APOBR: Apolipoprotein B47 receptor.
- Orexin Receptor 1.
- PIK3IP1 (PI3K-interacting protein 1) regulates PI3K catalytic subunit binding.
- TCF20: Transcription factor that recognizes PDGF-responsive element in the MMP3 promoter. Enhances JUN and SP1. Mutations associated with Autism.
- SKAP1: T cell adaptor protein.
- FAM109B (Family with sequence similarity 109 member B)
- MEF2C (MADS box transcription enhancer factor 2 C). Encodes MEF2 polypeptide C, which has DNA binding and trans-activation activities.
- NEGR1 (Neuronal growth regulator 1)
- ATP2A1-AS1 (ATP2A1 antisense RNA 1)
A genome-wide association study for extremely high intelligence (Zabaneh et al. 2017) #
- compared n=8185 controls with n=1247 individuals with IQ ~170.
- ADAM12 rs4962322, rs4962520 (C>T) and rs10794073. most associated with IQ… so is FNBP1L
Ashkenazi Genetics #
-
Natural history of Ashkenazi intelligence
- The two most important genetic disease clusters among the Ashkenazim are the sphingolipid storage disorders (Tay-Sachs, Gaucher, Niemann-Pick, and mucolipidosis type IV) and the disorders of DNA repair (BRCA1, BRCA2, Fanconi’s anemia type C, and Bloom syndrome)
-
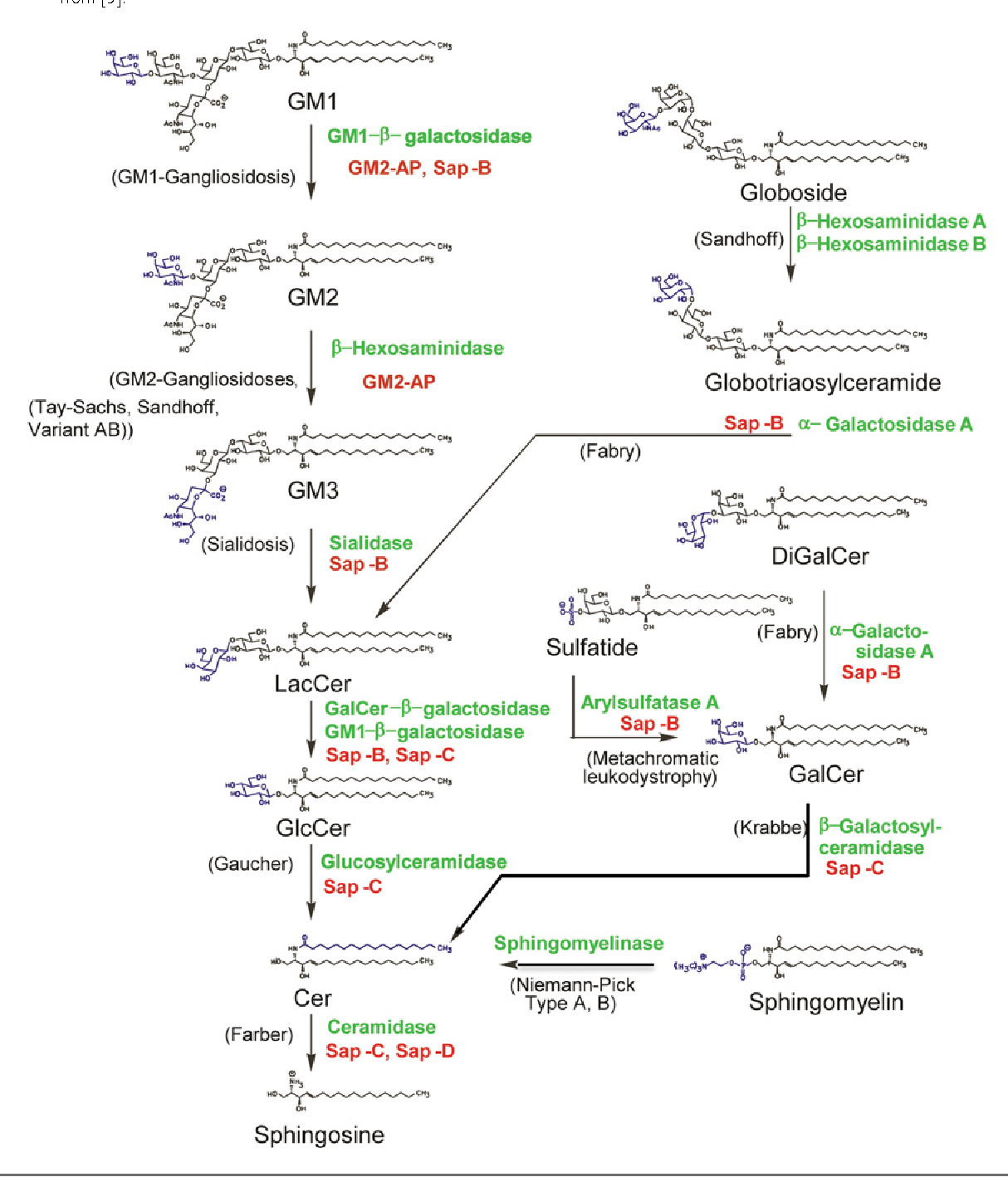
- The key datum is the effect of increased levels of the storage compounds. Glucosylceramide, the Gaucher storage compound, promotes axonal growth and branching: A regulatory role for sphingolipids in neuronal growth. Inhibition of sphingolipid synthesis and degradation have opposite effects on axonal branching
- In vitro, decreased glucosylceramide results in stunted neurones with short axons while an increase over normal levels (caused by chemically inhibiting Glucocerebrosidase) increases axon length and branching. There is a similar effect in Tay-Sachs: decreased levels of GM2 ganglioside inhibit dendrite growth, while an increase over normal levels causes a marked increase in dendritogenesis. ( Gangliosides as modulators of dendritogenesis in normal and storage disease-affected pyramidal neurons)
-
Tumor suppressor genes aren’t perfect, and will inhibit growth as a safeguard against cancer, killing and inhibiting growth of normal cells as collateral damage. It’s theorized that having less tumor-suppressor genes allows the brain to grow more. Ashkenazi Jews have many mutations in tumor-suppressor genes. No wonder.
-
Canavan disease: defective aspartocyclase, which breaks down N-Acetylaspartate resulting in massive neuronal cell death and mental retardation/death. NAA happens to also be a correlative of intelligence/creativity, and thus heterzygote carriers of Canavan’s disease have greater NAA, and probably greater intelligence.
-
A way to mimic spingolipid/ganglioside would be FIASMAs.
-
Mimicking the tumor-suppressor knockouts would probably be dangerous, but still doable. BRCA1, a commonly mutated gene in Ashkenazi Jews, works through p53, a tumor suppresor. Omigapil is a prime mimetic of BRCA1. Cancer risk though. Intranasal would be far better than oral.
-
Cholesterylester transfer protein (inhibiton)
-
NDST3 (why Einstein’s son had schizophrenia?) step in the production of Heparin.
Gwern: The Algernon Argument #
It’s a pretty short article outside of the numerous external links at the bottom of the page. https://www.gwern.net/docs/iq/index there’s plenty here, too.
Development #
-
- Words spoken at age 2 predicted ~20% of IQ variance at age 12 R
- Reading aloud leads to higher IQs in children R
-
Cognitive ability changes and dynamics of cortical thickness development in healthy children and adolescents Mean age 11.59 years. Assessed 2 years apart.
- No significant associations between changes in IQ measures and changes in cortical surface area were observed, whereas changes in FSIQ, PIQ, and VIQ were related to rates of cortical thinning, mainly in left frontal areas.
Giftnedness #
-
https://www.davidsongifted.org/gifted-blog/small-poppies-highly-gifted-children-in-the-early-years/
- The average age for a baby to utter their first word is about 12 months.
- The mean age at which 52 children of IQ 160+ uttered their first word was 9.1 months. 2 outliers removed and it’s 8.63. 16 months for sentences. 11 of them spoke their first meaningful word by 6 months.
- Robinson (1987) reports a young boy whose first utterance, at 20 months, was “Look! Squirrel eating birds’ food!”
- Gifted chilren learn mobility much earlier. Gross’s IQ 160+ subjects mean age of unsupported walking was 12.1 months - 3 months earlier than usual.
-
90% of them were reading before their 5th birthday. I suspect Latent Inhibition (curiosity) is a major aspect. I was thinking earlier if you remember all the shit you’ve been taught at school to this, like all the different types of clouds in 4th grade or whatever, your interest in these things must have sufficiently compensated for how inefficient these things are taught to just go in one ear and out the other.
- The average age for a baby to utter their first word is about 12 months.
-
Stimulation-seeking children, who might be seeking out and creating enriched environments for themselves, score higher on IQ tests and do better at school than less stimulation-seeking children (Raine et al. 2002).
Biological Models #
Network Neuroscience Theory of Human Intelligence #
https://www.cell.com/action/showPdf?pii=S1364-6613%2817%2930221-8 (Barbey, 2018)
- There is a correlation between lateral thinking and intelligence.
Interneuron Energy #
Cross frequency coupling #
- The brainweb: Phase synchronizaton and large-scale integration (On neurotyping, might make a page for this)
Working Memory #
Psychological Models #
g $\neq$ ‘general intelligence’. It’s really the “general factor”.
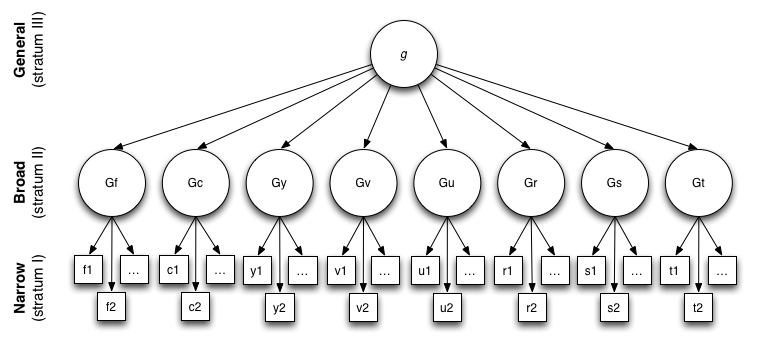
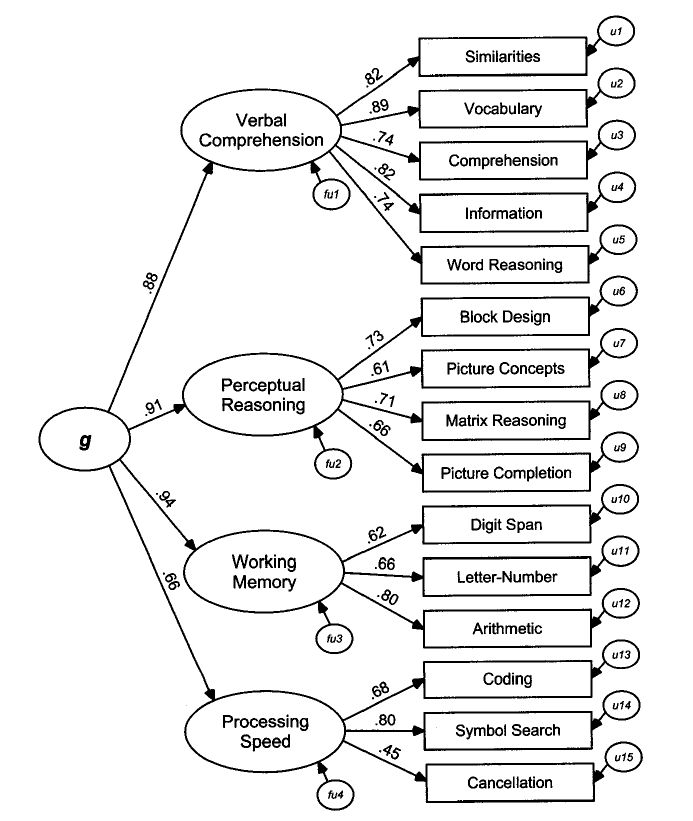
Cattel-Horn-Carroll theory #
Published in 1993. An integration of Cattel and Horn’s 1963 Gf/Gc theory and Carrol’s 1993 three-stratum theory.
 Makes sense that high WM phenotypes and the notion that sparser if finding relevant attractors is some kind of random search
g correlates with all these subfactors, and all the subfactors correlate ith each other.
Makes sense that high WM phenotypes and the notion that sparser if finding relevant attractors is some kind of random search
g correlates with all these subfactors, and all the subfactors correlate ith each other.
- Gf: Fluid intelligence
- Gc: Crystallized intelligence
- Gq: Quantitative reasoning
- Ability to comprehend quantitative concepts/relationships and manipulate numerical symbols
- Grw: Reading & writing ability
- Gsm: Short-term memory
- (Working memory; apprehend and use info within a few seconds)
- Glr: Long-term storage and retrieval
- Gv: Visual processing
- Ga: Auditory processing
- Gs: Processing speed
- Ability to perform automatic cognitive tasks, particularly when measured under pressure to maintain focused attention. (Measured in intervals of like 2-3 minutes)
- I thought this was IQ, bro?
- Gt: Decision/reaction time/speed (measured in <=1 second)
g-VPR model #
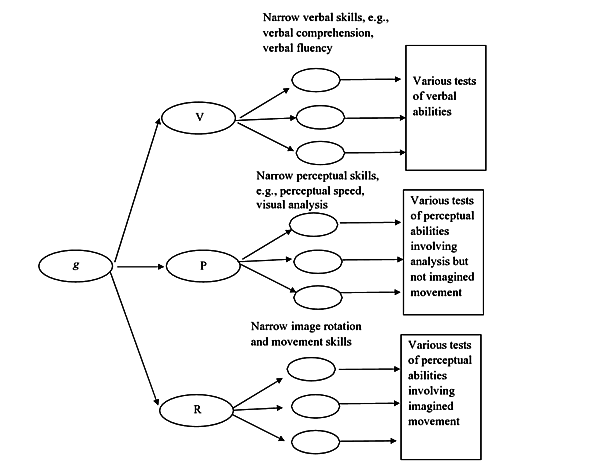 Published in 2005. Similar to the three-stratum theory (some regard it as superior to CHC).
There is g, and subitems of it are Verbal, Perceptual, and Rotation skills, and subitems of these are various narrow sub-skills for various tests of the ability to which they are subitems off.
Published in 2005. Similar to the three-stratum theory (some regard it as superior to CHC).
There is g, and subitems of it are Verbal, Perceptual, and Rotation skills, and subitems of these are various narrow sub-skills for various tests of the ability to which they are subitems off.
Birds #
- Driven by the need to stay light enough to fly, birds have scaled down their neurons to a level unmatched by any other group. Elephants have about 7,000 neurons per mg of brain tissue. Humans have about 25,000. Birds have up to 200,000. That means a small crow can have the same number of neurons as a pretty big monkey.
- https://tanyewwei.com/blog/pufa-birds-genetics/ Birds are insane. They thrive on a high PUFA diet yet the net effect is actually less PUFA in their (insanely powahful) Mitochondrial membranes.
- Alex Chen writes about this quite a bit.
- https://slatestarcodex.com/2019/03/25/neurons-and-intelligence-a-birdbrained-perspective/