Genetic Engineering
2022-07-30: Genetics reference:
- https://openwetware.org/wiki/Main_Page lab procedures
- https://openpcr.org/
- https://makezine.com/2016/07/18/arduino-powered-bioreactors-make-home-experimentation-affordable/
- https://makezine.com/2017/04/11/how-to-set-up-your-own-lab/
- https://yewtu.be/channel/UCV5vCi3jPJdURZwAOO_FNfQ
- https://biohackinfo.com/genetic-engineering/ some interesting genes for enhancement. I recognize a few and they’re definitely multifunctinoal
- On gods, pixies and humans: Biohacking and the genetic imaginary
-
https://diyhpl.us/wiki/
- Absolute gold mine: https://diyhpl.us/wiki/genetic-modifications/. Everything youuu need to know about Intelligence and other things in your Genome
Genetic Engineering/Biohacking #
- A plasmid is an extrachromosomal DNA molecule (thus is not part of the genome) within a cell that can independently replicate. Thus, when you get a plasmid it’s probably already on bacteria so that you only have to do the insertion once.
- So, you send plasmid-containing bacteria into your body and make sure it penetrates (relevant?) cells.
- Get them on addgene.org or something
- So, you send plasmid-containing bacteria into your body and make sure it penetrates (relevant?) cells.
- Adeno Associated Virus is just some useless virus that most people have in their tissues. It can’t replicate without actual adenoviruses.
- Plasmids locate into the nucleus to be translated to RNA.
-
Stem cell-derived clade F AAVs mediate high-efficiency homologous recombination-based genome editing
- AAVHSC: Hematopoietic stem cell-derived adeno-associated virus vector.
- Zayner says this has an efficiency of >~50%, while CRISPR is only ~1%.
- So before CRISPR, there was ZFN (zinc finger nucleases) and TALENs (transcrpition activator-like effector nucleases). The first knockout mouse was in 1989, which won the guys a 2007 nobel prize.
- There’s an imune system component, I believe, to rejecting newly-inserted DNA/their encoded proteins, especially for haematological genes.
- Identification of Pre-Existing Adaptive Immunity to Cas9 Proteins in Humans
- On the flip side, CRISPR can be used to suppress the imune response to stem cell transplants.
- Base/BASE editing changes SNPs (or in this case, SNVs (variants)).
-
- The risk for only a fraction of the cells in a targeted organism, area, etc. being successfully edited is referred to as mosaicism.
- Predicting base editing outcomes with an attention-based deep learning algorithm trained on high-throughput target library screens (Aug 2021)
- The first ever in-body gene edcit was 13 Nov 2017 for a man with Hunter syndrome, replacing his gene with a healthy one. However, one month before this, Zayner did his stunt as possibly the first ever gene edit! Things started picking up speed in 2020: AAV gene therapy for Tay-Sachs disease (Feb 2022)
-
Significantly Enhancing Adult Intelligence With Gene Editing May Be Possible
-
Cytosine and adenine base editing of the brain, liver, retina, heart and skeletal muscle of mice via adeno-associated viruses
- Optimized dual AAVs enable in vivo base editing at therapeutically relevant efficiencies and dosages in the mouse brain (up to 59% of unsorted cortical tissue), liver (38%), retina (38%), heart (20%) and skeletal muscle (9%).
-
Cytosine and adenine base editing of the brain, liver, retina, heart and skeletal muscle of mice via adeno-associated viruses
CRISPR #
- CRISPR: clustered regulatory interspaced short palindromic repeats. Just a categorization/family of DNA sequences in bacteria, archaea, prokaryotes. It’s part of the human genome, and is part of the prokaryotic adaptive immune system.
- How bacteria get spacers from invaders
-
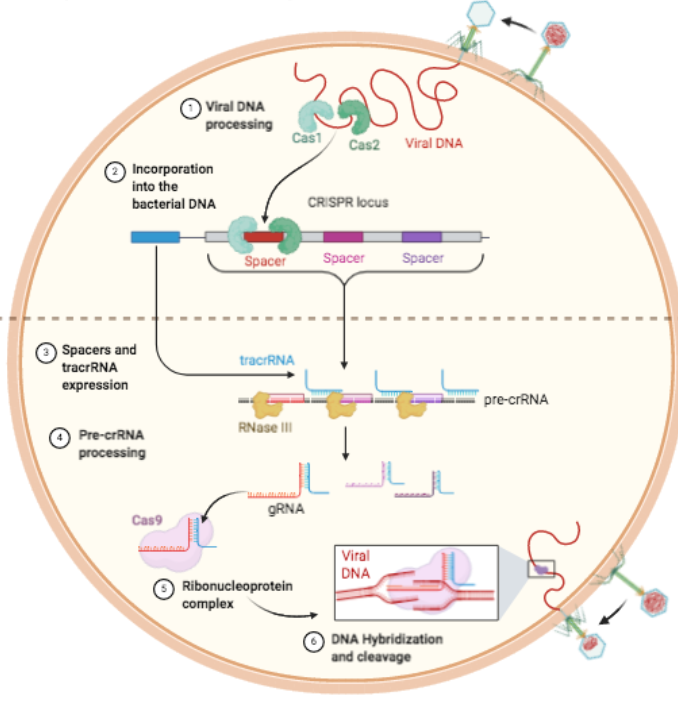 Between the palindromic repeats are ‘spacers’, which are foreign DNA associating from mobile genetic elements which is either genetic material from another species, or move around within the genome, e.g. plasmids, bacteriophages, and transposons.
Between the palindromic repeats are ‘spacers’, which are foreign DNA associating from mobile genetic elements which is either genetic material from another species, or move around within the genome, e.g. plasmids, bacteriophages, and transposons.
- Cas cleaves the viral DNA, and is also responsible for then integrating it into the CRISPR array as a spacer(s). The part of the invading DNA that will form the spacer is known as the protospacer.
- The CRISPR array is then eventually transcribed to pre-crRNA whereby trans-activating crRNA (
- Then, Cas, e.g. Cas9 (found in Streptococcus pyogenes (so SpCas9)) associates with guide RNA to form the gRNA-Cas9 complex, which then is activated to cleave its the gRNA’s associated DNA as part of the immune response.
- Thus, By delivering Cas9 complexed with a synthetic gRNA into a cell, the cell’s genome can be cut at a desired location, allowing existing genes to be removed and/or new ones added in vivo.
- Relevant are PAMs (protospacer adjacent motifs), which is required for cleavage via Cas9. Without it, the cell’s own CRISPR locus could get cleaved, no?! A few base pairs upstream is where Cas cuts:
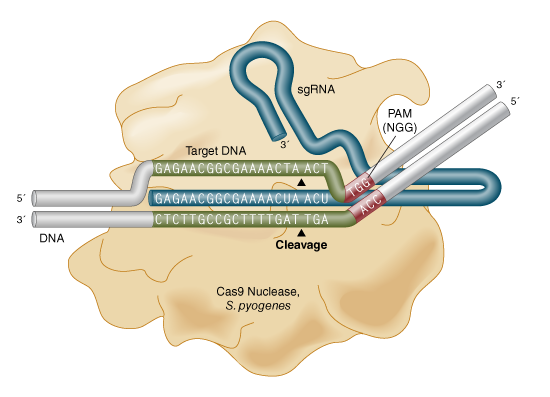
- Not really depicted, but Cas9 has two cleavage domains for the target DNA strands.
- Soo, you’ll need PAMs in your genome, too. SpCas9 cuts upstream of 5’-NGG-3’ where N is any nucleotide base. SaCas9 cuts 5’-NNGRR(N)-3’.
- sgRNA = single/synthetic guide RNA (also a name for subgenomic RNA). It is produced by synthetic linkage of pre-crRNA and tracrRNA that constitute gRNA. The gRNA can be for any 20-nucleotide sequence.
- It can also be made in vitro/vivo with some kind of DNA template but afaik it’s not found naturally.
-
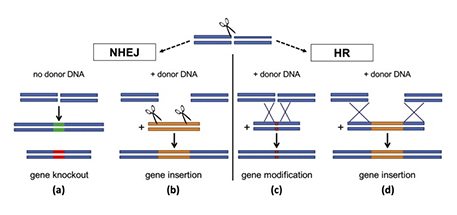
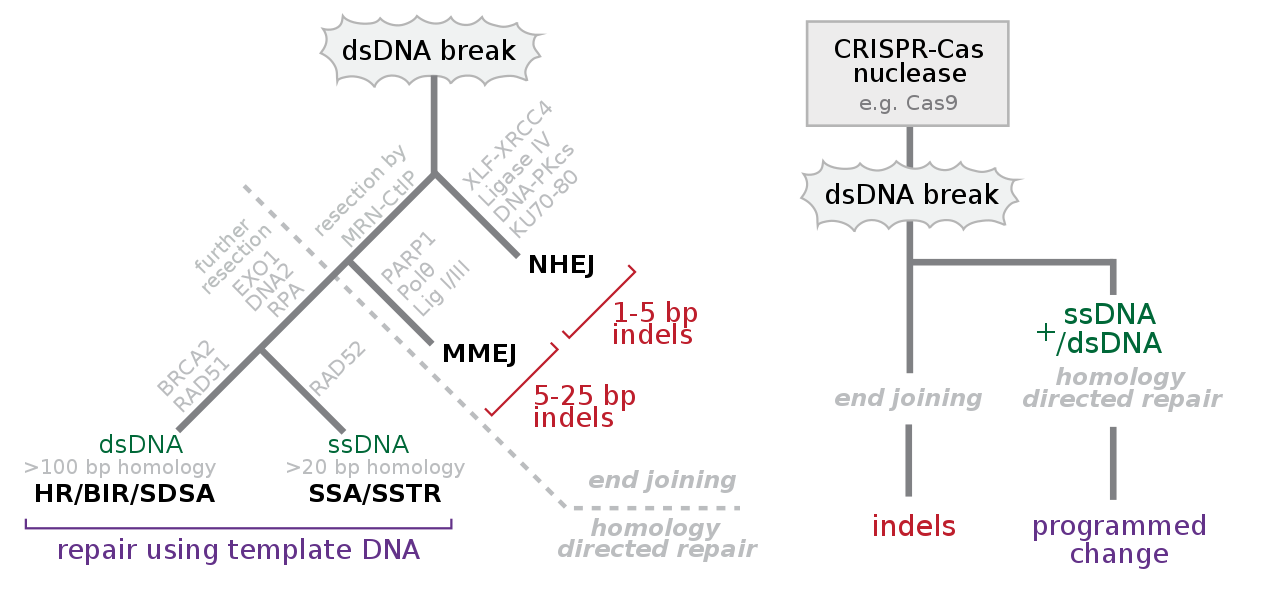
- Non-Homologous End Joining (NHEJ): indels; deletion or insertion. After cleavage, dsDNA (double-stranded broken) can get left repairing normally, which often has errors I believe, hence causing knockout, or one can artificially supply an entirely new sequence to replace it with which is achieved with donor DNA.
- HDR (homology-directed repair) is not an indel; a selling point is that it does not change the size of DNA. The template has homologous DNA sequences to the surrounding site of cleavage, with new DNA betwixt.
Editing #
- Design your sgRNA with: synthego, crispor, crispr-era, cas-offfinder, off-spotter, E-CRISP, https://www.addgene.org/crispr/reference/grna-sequence/, https://www.atum.bio/eCommerce/cas9/, http://chopchop.cbu.uib.no/
- After obtaining desired sgRNA, you have to purify it from bacteria. Mix with polyethylenimine(PEI) at a ratio of about 1μg DNA to 10μg PEI and inject >20μg of DNA. Injections will most likely need to be done multiple times to engineer enough cells to have an effect.
- There are certain loci in the genome to be targeted for ubiquitous, constitutive expression, like ROSA26 (mouse chromosome 6… has its equivalent somewhere) or AAVS1 (human chromosome 19).
- When targeting your gRNA, 40-60% GC content is desirable, and chromatin accessibility is important.